Data
for Democracy, Fall 2024
Andy Lyons

tidyCensus
Responsibility of counties and states to collect and report voting precinct boundaries and turnout
Availability, quality, and format varies a lot from state-to-state
Organizations & initiatives that collect and publish election data:
state election boards
Redistricting Data Hub - project under New Venture Fund (NVF), implemented by HaystaqDNA
MGGG Redistricting Lab - Tufts University, Cornell
https://redistrictingdatahub.org/
The nonpartisan Redistricting Data Hub provides individuals, good government organizations, and community groups the data, resources, and knowledge to participate effectively in the redistricting process.




These packages allow you to:
Example:
sfTo import GIS data with sf, you have to specify a
source and layer.
The Source can be a:
folder: "./gis_data"
geodatabase (which is really a folder):
"./gis_data/yose_roads.gdb"
file: "trail_heads.kml",
"cell_towers.geojson"
database connection string:
"PG:dbname=postgis"
In sf functions, the argument where you provide the
source is often named dsn (data source
name)
The Layer can be a:
Shapefile name (minus the shp extension)
a named layer in the database / file
The two main functions for importing vector data are:
st_layers(source) - returns the names of available layers in a source
st_read(source, layer) - import into R
Most of the functions in the sf package start with
st_, which stands for ‘space time’, and
matches the names of similar functions in PostGIS.
To view the metadata of a layer before bringing it into R, use
rgdal::ogrInfo()
\*.shp,
\*.shx, \*.prj, \*.dbf,
etc.
st_read(dsn, layer)
dsn - directory, or shp filename
layer - shp filename (minus .shp), or omitted
library(sf)
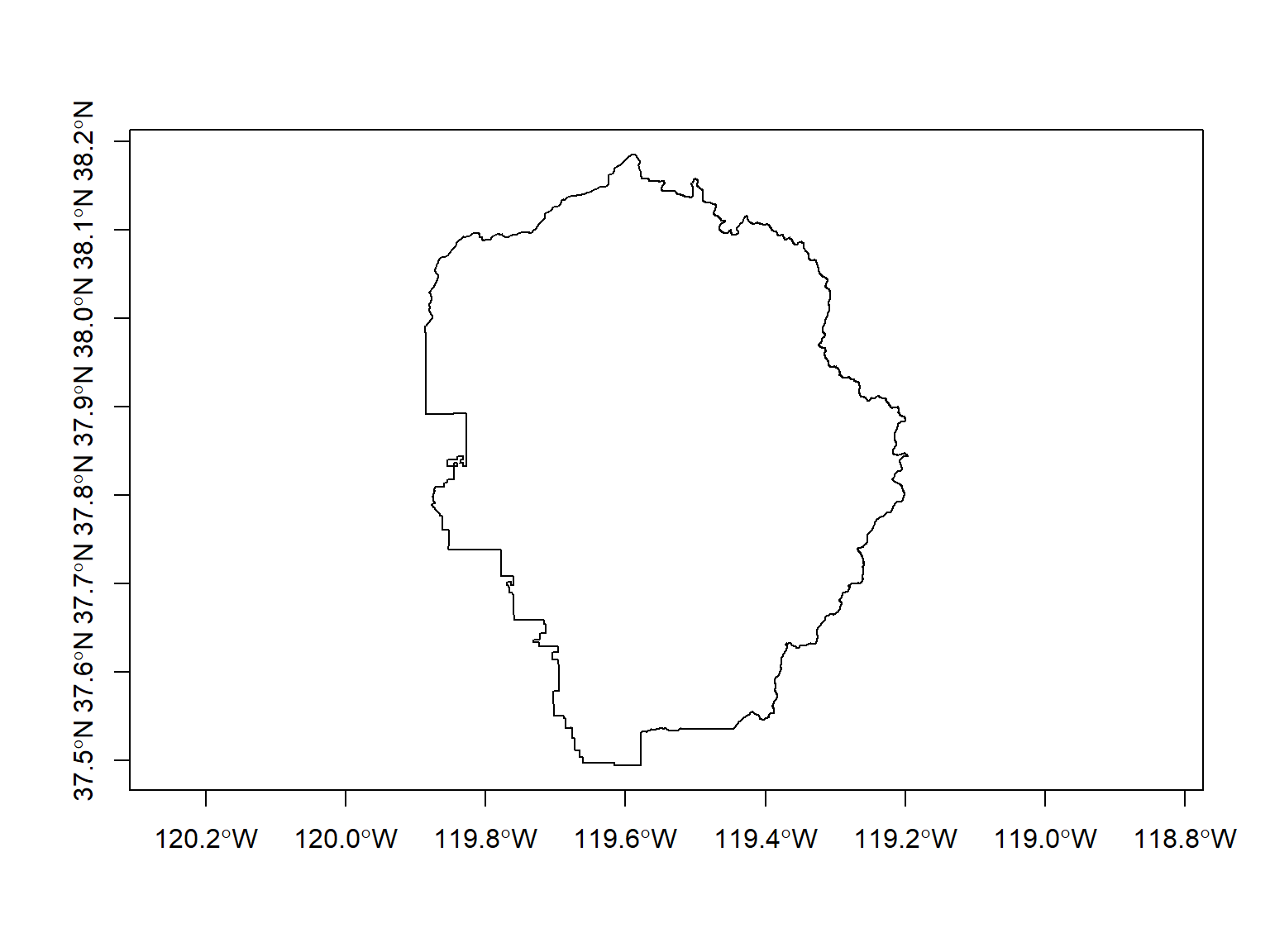
yose_bnd_ll <- st_read(dsn="./data", layer="yose_boundary")
## This would also work:
## yose_bnd_ll <- st_read(dsn="./data/yose_boundary.shp")
View contents:
## Simple feature collection with 1 feature and 11 fields
## Geometry type: POLYGON
## Dimension: XY
## Bounding box: xmin: -119.8864 ymin: 37.4947 xmax: -119.1964 ymax: 38.18515
## Geodetic CRS: North_American_Datum_1983
## UNIT_CODE
## 1 YOSE
## GIS_NOTES
## 1 Lands - http://landsnet.nps.gov/tractsnet/documents/YOSE/METADATA/yose_metadata.xml
## UNIT_NAME DATE_EDIT STATE REGION GNIS_ID UNIT_TYPE
## 1 Yosemite National Park 2016-01-27 CA PW 255923 National Park
## CREATED_BY METADATA PARKNAME
## 1 Lands http://nrdata.nps.gov/programs/Lands/YOSE_METADATA.xml Yosemite
## geometry
## 1 POLYGON ((-119.8456 37.8327...
Plot:
| join data frames on a column | left_join(), right_join(), inner_join() |
| stack data frames | bind_rows() |
To join two data frames based on a common column, you can use:
left_join(x, y, by)
where x and y are data frames, and by is the name of a column they have in common.
If there is only one column in common, and if it has the same name in both data frames, you can omit the by argument.
If the common column is named differently in the two data frames, you can deal with that by passing a named vector as the by argument. See below.
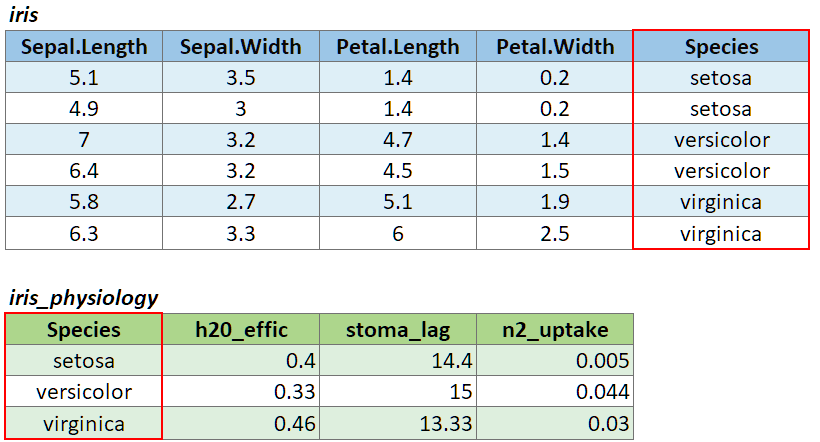
To illustrate a table join, we’ll first import a csv with some fake data about the genetics of different iris species:
# Create a data frame with additional info about the three IRIS species
iris_genetics <- data.frame(Species=c("setosa", "versicolor", "virginica"),
num_genes = c(42000, 41000, 43000),
prp_alles_recessive = c(0.8, 0.76, 0.65))
iris_genetics## Species num_genes prp_alles_recessive
## 1 setosa 42000 0.80
## 2 versicolor 41000 0.76
## 3 virginica 43000 0.65We can join these additional columns to the iris data frame with
left_join():
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species num_genes
## 1 5.1 3.5 1.4 0.2 setosa 42000
## 2 4.9 3.0 1.4 0.2 setosa 42000
## 3 4.7 3.2 1.3 0.2 setosa 42000
## 4 4.6 3.1 1.5 0.2 setosa 42000
## 5 5.0 3.6 1.4 0.2 setosa 42000
## 6 5.4 3.9 1.7 0.4 setosa 42000
## 7 4.6 3.4 1.4 0.3 setosa 42000
## 8 5.0 3.4 1.5 0.2 setosa 42000
## 9 4.4 2.9 1.4 0.2 setosa 42000
## 10 4.9 3.1 1.5 0.1 setosa 42000
## prp_alles_recessive
## 1 0.8
## 2 0.8
## 3 0.8
## 4 0.8
## 5 0.8
## 6 0.8
## 7 0.8
## 8 0.8
## 9 0.8
## 10 0.8
If you need to join tables on multiple columns, add additional column
names to the by argument.
Join columns must be the same data type (i.e., both numeric or both character).
There are several variants of left_join(), the most
common being right_join() and
inner_join(). See help for details.
If the join column is named differently in the two tables, you can
pass a named character vector as the by argument. A named
vector is a vector whose elements have been assigned names. You can
construct a named vector with c().
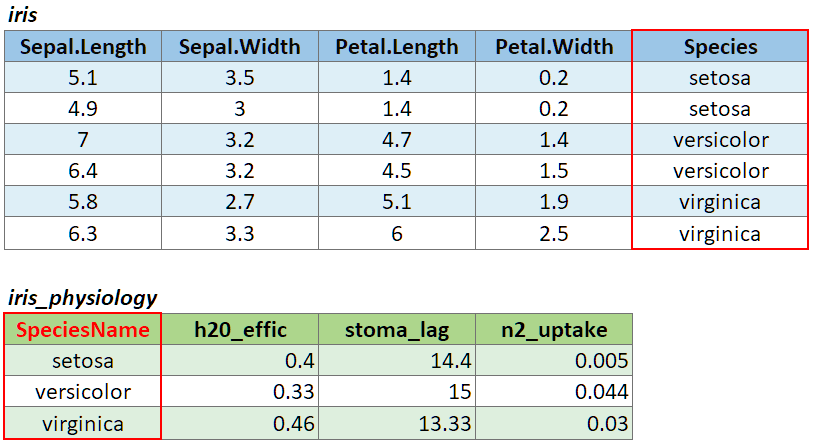
For example if the join column was named ‘SpeciesName’ in x, and just ‘Species’ in y, your expression would be:
left_join(x, y, by = c("SpeciesName" = "Species"))

Reshaping data includes:
The go-to Tidyverse package for reshaping data frames is tidyr
In this exercise, we will: